1. Project Title: Triclustering algorithms for Three-Way Medical Data Analysis: Targetting efficient algorithms with significant solutions
2. Area of knowledge: Physical Sciences, Mathematics and Engineering Panel
3. Group of disciplines: Theoretical and Applied Mathematics, Computer Sciences and IT
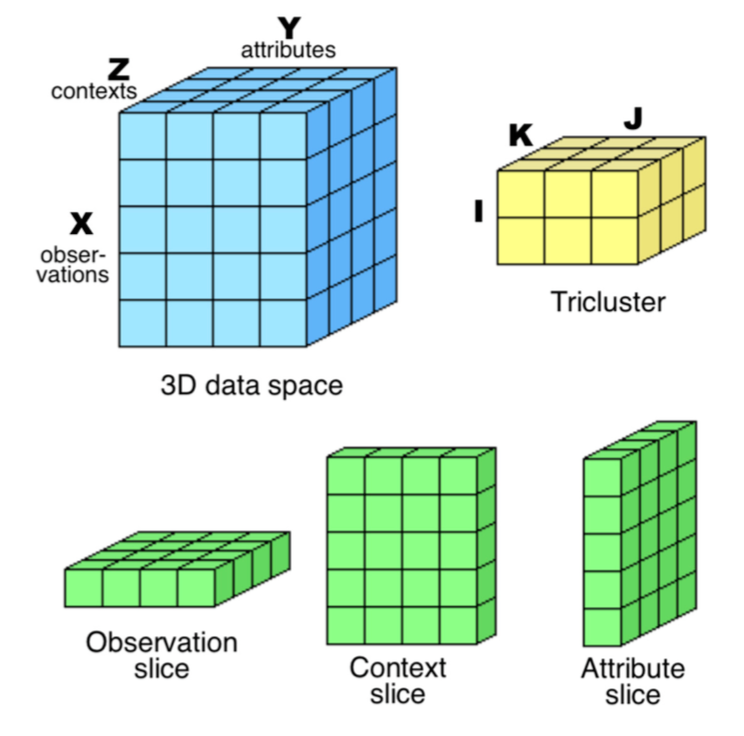
4. Research project: Three-way (3W) data, also referred to as tridiac, three-dimensional, tensorial, or cubic, are increasingly prevalent. In biomedical domains, the periodic profiling of entities and clinical monitoring of individuals give rise to multivariate time series. For example, the analysis of patient-record-time data is essential to understand the complex physiological processes underlying disease progression and responses to stimuli, drugs, or therapy. In social domains, understanding consumerism, web usage, work perfor-mance, and social activity has been done using individual-feature-time data. Financial decisions can be supported by the analysis of stock-ratio-time data or society-society-trade data. The aforementioned forms of 3W data are generally referred to as observation-attribute-context data, and the term object is used to indistinctly denote an observation, attribute, or context. Due to the complexity of biomedi-cal and social 3W data analysis, a natural first step is to identify potentially relevant subsets of objects. Standard clustering can be used to group objects and is usually performed globally using all the attrib-utes, an undesirable restriction in 3W data with locally correlated values. Subspace clustering was orig-inally proposed to address this issue and, in the context of 2W data, it is known as biclustering. In the context of 3W data, subspace clustering is termed triclustering. Given 3W data, triclustering aims at finding subsets of observations, attributes, and contexts (triclusters) satisfying certain homogeneity and statistical significance criteria. Triclustering can be applied to unravel regulatory modules, disease patterns, communities with shared behavior, thus being key to study complex biological, individual, and societal systems. This fact justifies the increasing attention given to triclustering, an emerging re-search topic in data analysis and machine learning (Henriques and Madeira, 2018, https://dl.acm.org/citation.cfm?doid=3271482.3195833).
5. Job position description: Despite its relevance, triclustering still faces major challenges: computational efficiency; homogeneity, as coherence criteria need to be designed for the target application; avoidance of biases in favor of one or more dimensions; robustness to varying types (and levels) of noise and missing data, common in real-world data; statistical significance. In this scenario, the candidate will embrace 2 major challenges: 1) computational efficiency, key to promote triclustering application to real-world large-scale 3W data; and 2) statistical significance, key to quantify the level of confidence on results. The work will focus 3D Medical Data Analysis, with 3 high impact triclustering applications: 1) multivariate physiological signal (individual-feature-time signal data) analysis, where triclusters can capture coherent physiological responses for a group of individuals; 2) neuroimaging data analysis, where triclusters can capture hemodynamic response functions and connectivity between brain regions; and 3) clinical records analysis, where triclusters correspond to groups of patients with correlated clinical features along time. In this context, a large private dataset of genotype-phenotype data and clinical temporal data collected by national FCT project NEUROCLINOMICS2 and European JPND project OnWebDuals, concerning the study of patients with Amyotrophic Lateral Sclerosis, together with public data from brain map will be used (https://portal.brain-map.org) will be used. The candidate will integrate a highly multidisciplinary research team, gathering unique computational expertise in data science/machine learning applied to neurodegenerative diseases with strong and long term collaborations with clinicians and a renowned neurologist/researcher in ALS. Candidates should have an MSc in Computer Science, Bioinformatics, Data Science or related areas, should be proficient in programming and English. Multidisciplinary backgrounds are welcomed.
6. Group leader:
The work will be supervised by Sara C. Madeira, Associate Professor at the Department of Informatics, Faculty of Sciences (FCUL), University of Lisbon, where she coordinates the Msc in Data Science and teaches courses on Data Mining, Machine Learning, Foundations of Data Science, and Intelligent Systems. She is also senior Researcher at LASIGE, where she coordinate the Data and Systems Intelligence Research Line of Excellence (DSI) and is also a member of the Health and Biomedical Informatics (HBI). The candidate will be work in LASIGE integrated in DSI and HBI Research Line of Excellence. Full name: Sara Alexandra Cordeiro Madeira Email: sacmadeira@ciencias.ulisboa.pt Google scholar profile: https://scholar.google.com/citations?hl=en&user=rpaGL7AAAAAJ&view_op=list_works&sortby=pubdate